Deplancke lab – Laboratory of Systems Biology and Genetics

©EPFL, Deplancke's lab
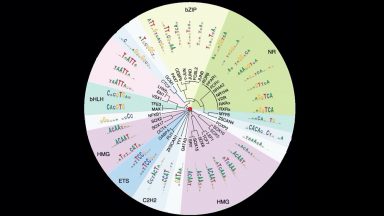
The central research focus of the Laboratory of Systems Biology and Genetics (LSBG) is to understand genome organization, regulation, and variation and is broadly partitioned according to three major research pillars:
Adipo
In the “Adipo” pillar, we are characterizing the regulatory mechanisms driving mesenchymal stromal cell heterogeneity and function in adipose biology and other metabolically relevant processes.

Geno
In the “Geno” pillar, we are examining how variation in the regulatory code affects molecular and organismal diversity.

Techno
In the “Techo” pillar, we are developing novel microfluidic, high-throughput sequencing, single cell omic, and computational approaches, which are used to address biological questions in the other two pillars.
Latest news
04.07.2023
Prof. Bart Deplancke elected EMBO member!
The European Molecular Biology Organization (EMBO) has elected 69 new life scientists to membership, joining more than 1800 of the best researchers in Europe and the world. Among the new members is Professor Bart Deplancke at EPFL’s Institute of Bioengineering.

22.08.2022
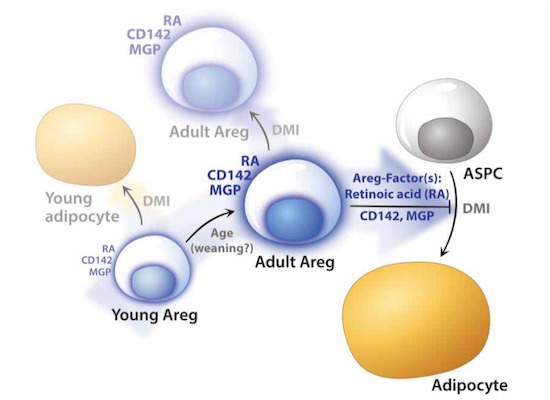
A recent profiling study by Magda Zachara, Pernille Rainer, Horia Hashimi et al. reports molecular and functional properties of an ASPC subpopulation termed “adipogenesis regulator” (Areg) cells, revealing unexpected paracrine-mediated adipogenic inhibition adjusted during lifetime. The article: “Mammalian adipogenesis regulator (Areg) cells use retinoic acid signalling to be non- and anti-adipogenic in age-dependent manner.” appeared in The EMBO Journal.
17.08.2022
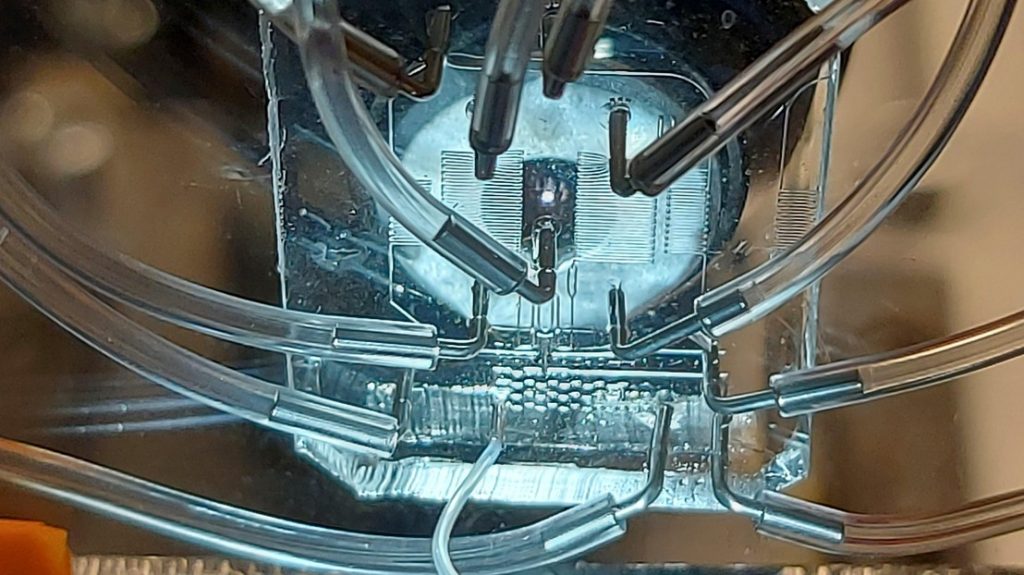
A new technique keeps cells alive during extraction to track the activity of thousands of genes across time. The breakthrough method is called Live-seq and was jointly developed by scientists from the groups of professors Bart Deplancke at EPFL and Julia Vorholt at ETH Zurich.
The work published in Nature is also highlighted by over 30 news outlets such as preLights (Preprints selected by the Biology Community); Le Monde (French), German National Radio (German), and Genetic Engineering & Biotechnology. Its Altmetric score has already exceeded 900.

15.06.2022
Professors Bart Deplancke awarded Advanced Grant from the Swiss National Science Foundation.
SinPhonies project description (Deplancke)
The ability to identify which genes are expressed in a single cell, called single cell transcriptomics (scRNA-seq), is revolutionizing biology by allowing us to address fundamental problems with unprecedented resolution, such as how cells develop into a plastic tissue or how they trigger disease. However, several major technological limitations of scRNA-seq remain.
The project, named SinPhonies for Single cell Phenomic technologies, aims to design two autonomous but synergistic technologies that each address a key limitation to advance single cell phenomena coupled to scRNA-seq.

28.04.2022
EPFL Professor Bart Deplancke awarded the Leenaards Science Prize with Dr Jeroen Geurts (CHUV) for a joint research project on obesity and osteoarthritis.
Official press release from Leenaards
14.02.2022
Bioengineers in Deplancke’s group have found a way to radically increase the efficiency of single-cell RNA-sequencing, a powerful tool that can “read” the genetic profile of an individual cell. Published in Nature Methods, the method is dubbed “DisCo” for “deterministic, mRNA-capture bead and cell co-encapsulation dropleting system”.
Our creations / innovations / inventions

Catalyze4life
Catalyze4life’s mission is to support translational projects and to educate to innovate

Junior European Drosophilia Investigators (JEDI)
The JEDIs are early career scientists working on Drosophila melanogaster in Europe.
Contact
Office Number: SV 3823
Administration: Joanna Rusnok
Office Number: SV 3515